Proteome - SMDBP000047 ( P07339 )
Description
- IDSMDBP000047
- NameCathepsin D (EC 3.4.23.5) [Cleaved into: Cathepsin D light chain; Cathepsin D heavy chain]
- OrganismHomo sapiens (Human)
- Gene SymbolCTSD
- Gene SynonymsCLN10; CPSD; HEL-S-130P
- ChromosomeGRCh38 chr11:1752755-1763927; hg19 chr11:1773985-1785157
- Gene Sequence
AACTGCGGCGTCATCCCGGCTATAAGCGCACGGCCTCGGCGACCCTCTCCGACCCGGCCGCCGCCGCCATGCAGCCCTCCAGCCTTCTGCCGCTCGCCCTCTGCCTGCTGGCTGCACCCGCCTCCGCGCTCGTCAGGATCCCGCTGCACAAGTTCACGTCCATCCGCCGGACCATGTCGGAGGTTGGGGG Show more...
- Protein Sequence
MQPSSLLPLALCLLAAPASALVRIPLHKFTSIRRTMSEVGGSVEDLIAKGPVSKYSQAVPAVTEGPIPEVLKNYMDAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIHHKYNSDKSSTYVKNGTSFDIHYGSGSLSGYLSQDTVSVPCQSASSASALGGVKVERQVFGEATKQPGITF Show more...
- Protein Length412
- Mass44552.0
External Links
- UniprotP07339
- HGNC2529
- HPAENSG00000117984
- GeneCardsCTSD
Expression
- Abundance
Export
Loci
- Sperm LociPrincipal piece
Sperm Loci
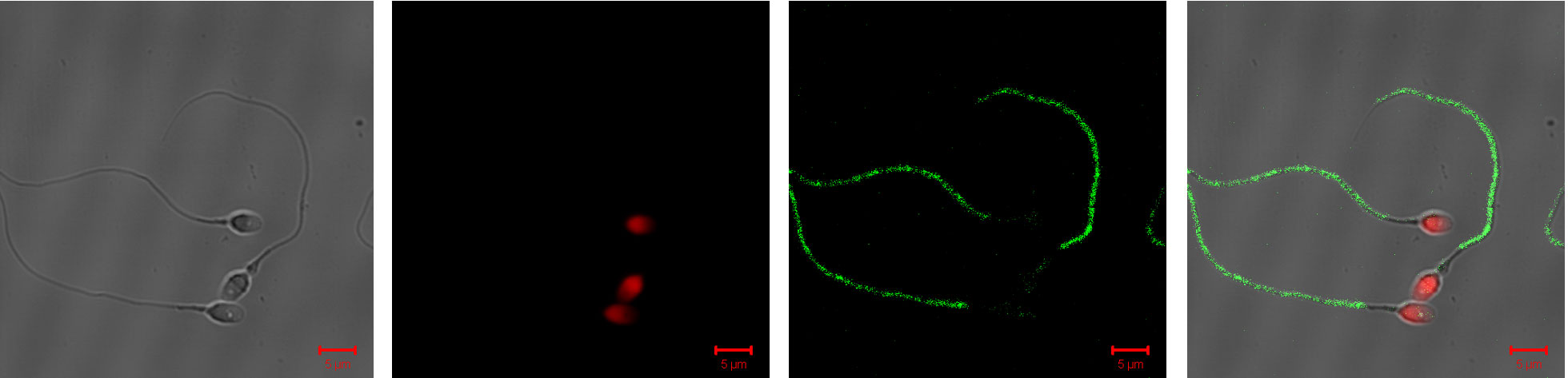
Immunocytochemistry of washed motile ejaculated human spermatozoa incubated with antibodies against epididymal secretory proteins and testicular proteins (right panels, merged micrographs of fluorescence staining). Red staining (by PI) indicates the nuclei and green staining (by FITC) indicates the location of HEL-S-130P on spermatozoa. Each bar represents 5 um.
Epididymis and Testis Loci

Immunohistochemical location of sperm-located and nonlocated proteins in tissue sections. proteins located in different regions (left panels, Caput; middle left panels, Corpus; middle right panels, Cauda; right panels, Testis) in different tissue compartments.1, negative control, preimmune rabbit serum; 2, HEL-S-181mP, in the epididymal epithelium , on microvilli and the testis of all four regions; 3, HEL-S-100n, in the epididymal epithelium , on microvilli and the testis of all four regions. Each bar represents 200 um.
Biological Properties
- General FunctionAcid protease active in intracellular protein breakdown. Plays a role in APP processing following cleavage and activation by ADAM30 which leads to APP degradation (PubMed:27333034). Involved in the pathogenesis of several diseases such as breast cancer and possibly Alzheimer disease. {ECO:0000269|PubMed:27333034}.
- GO – Biological Processantigen processing and presentation of exogenous peptide antigen via MHC class II [GO:0019886]; lipoprotein catabolic process [GO:0042159]; positive regulation of apoptotic process [GO:0043065]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280]; proteolysis [GO:0006508]; regulation of establishment of protein localization [GO:0070201]
- GO – Cellular Componentcollagen-containing extracellular matrix [GO:0062023]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; ficolin-1-rich granule lumen [GO:1904813]; lysosomal lumen [GO:0043202]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; melanosome [GO:0042470]; membrane raft [GO:0045121]; specific granule lumen [GO:0035580]; tertiary granule lumen [GO:1904724]
- GO – Molecular Functionaspartic-type endopeptidase activity [GO:0004190]; aspartic-type peptidase activity [GO:0070001]; cysteine-type endopeptidase activity [GO:0004197]; peptidase activity [GO:0008233]